×
![]()
neos-4343293-stony: Instance-to-Instance Comparison Results
| Type: | Instance |
| Submitter: | Jeff Linderoth |
| Description: | (None provided) |
| MIPLIB Entry |
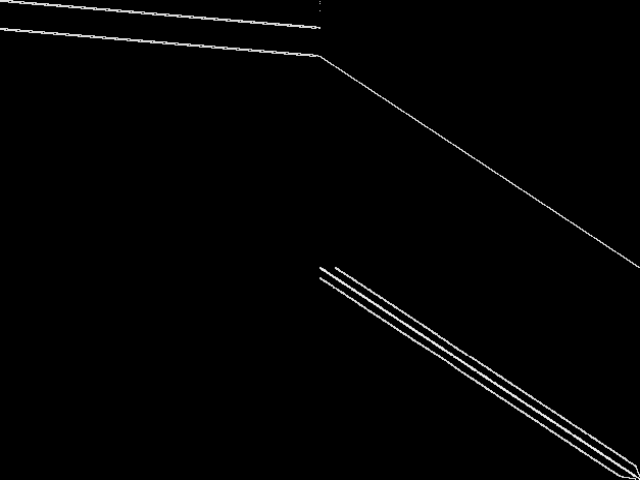
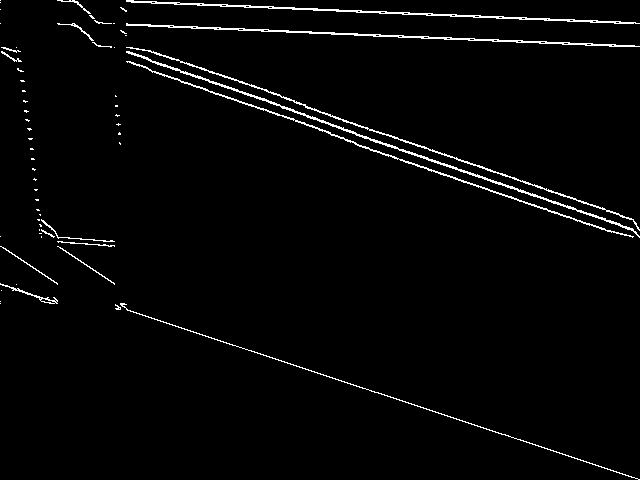
Parent Instance (neos-4343293-stony)
All other instances below were be compared against this "query" instance.  |
 |
 |
 |
 |
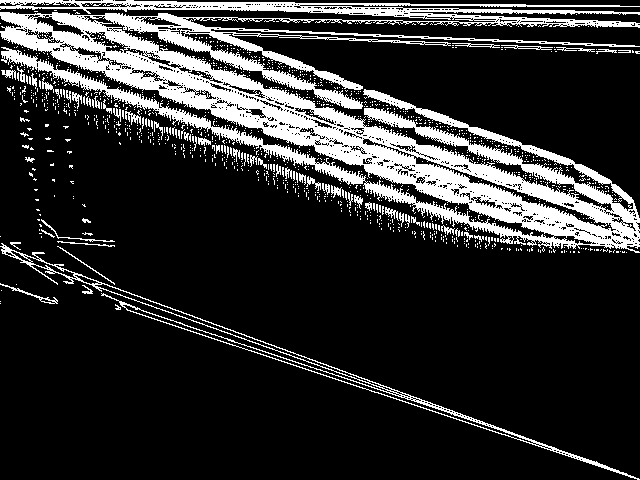 |
|
Raw
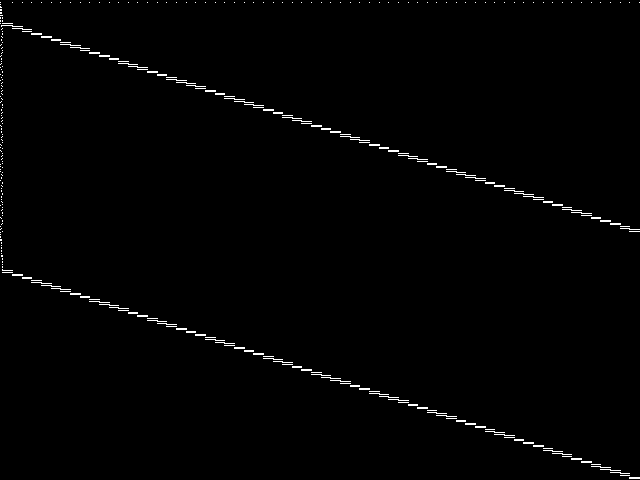
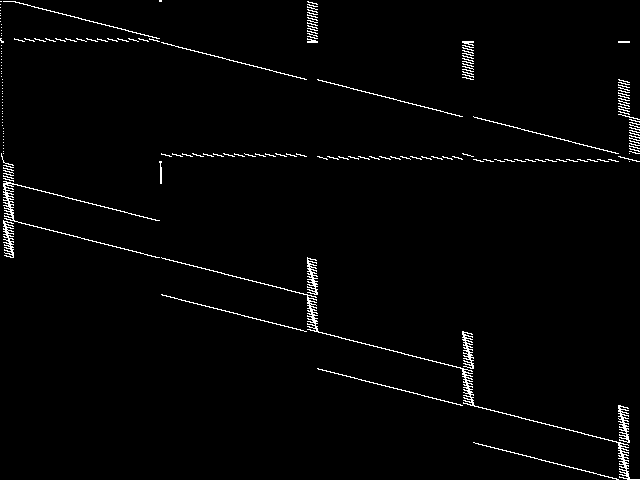
This is the CCM image before the decomposition procedure has been applied.
|
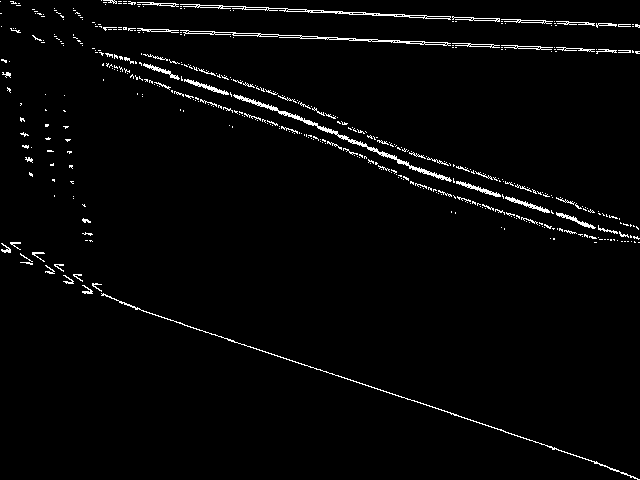
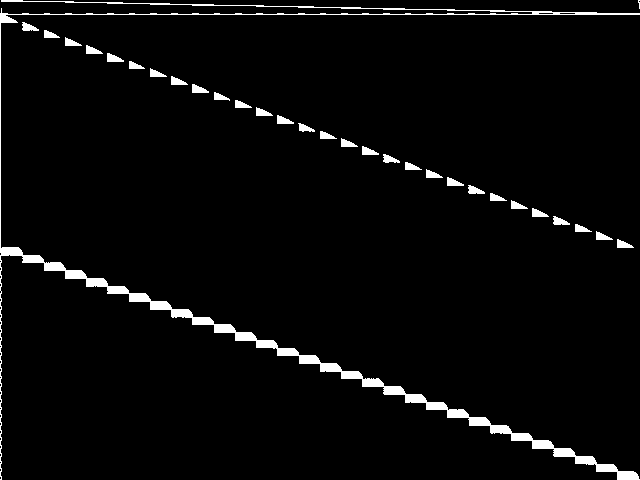
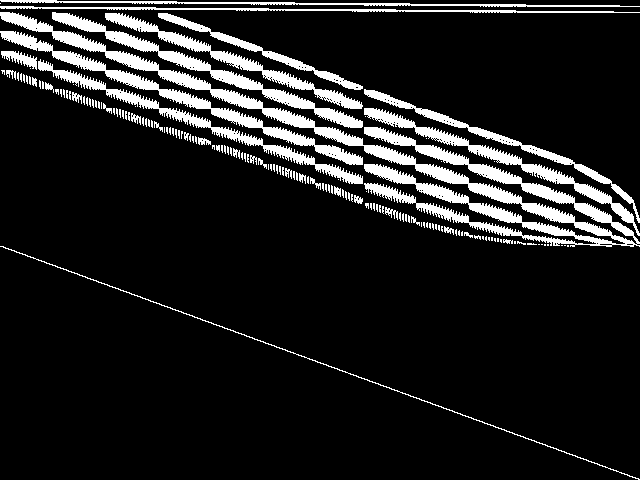
Decomposed
This is the CCM image after a decomposition procedure has been applied. This is the image used by the MIC's image-based comparisons for this query instance.
|
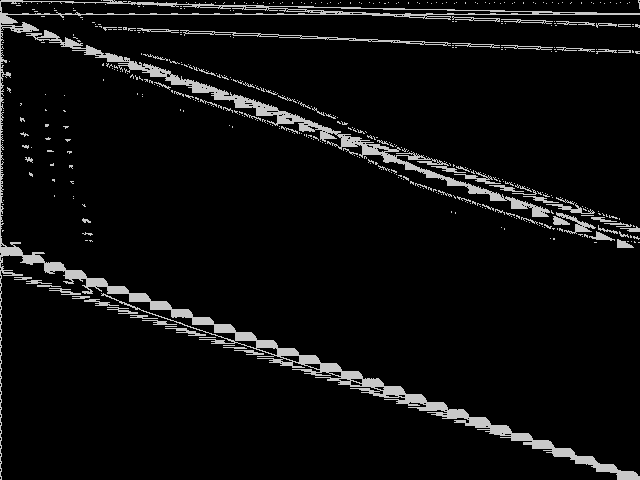
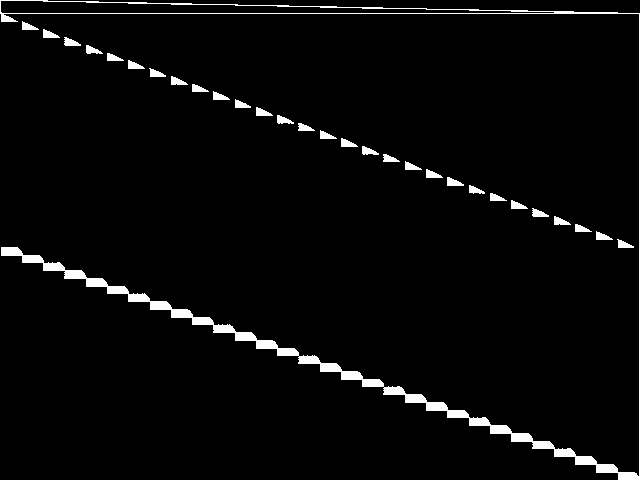
Composite of MIC Top 5
Composite of the five decomposed CCM images from the MIC Top 5.
|
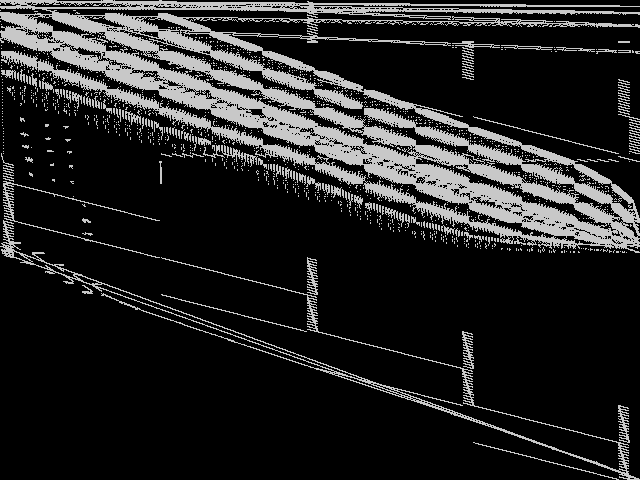
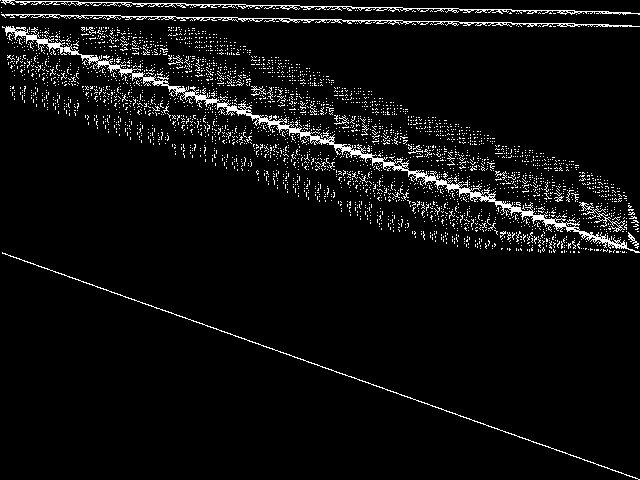
Composite of MIPLIB Top 5
Composite of the five decomposed CCM images from the MIPLIB Top 5.
|
Model Group Composite Image
Composite of the decomposed CCM images for every instance in the same model group as this query.
|
MIC Top 5 Instances
These are the 5 decomposed CCM images that are most similar to decomposed CCM image for the the query instance, according to the ISS metric.  |
Decomposed
These decomposed images were created by GCG.
|
 |
 |
 |
 |
 |
| Name | neos-4393408-tinui [MIPLIB] | proteindesign122trx11p8 [MIPLIB] | proteindesign121pgb11p9 [MIPLIB] | proteindesign121hz512p9 [MIPLIB] | supportcase43 [MIPLIB] | |
|
Rank / ISS
The image-based structural similarity (ISS) metric measures the Euclidean distance between the image-based feature vectors for the query instance and all other instances. A smaller ISS value indicates greater similarity.
|
1 / 0.819 | 2 / 0.991 | 3 / 0.992 | 4 / 0.996 | 5 / 1.009 | |
|
Raw
These images represent the CCM images in their raw forms (before any decomposition was applied) for the MIC top 5.
|
 |
 |
 |
 |
 |
MIPLIB Top 5 Instances
These are the 5 instances that are most closely related to the query instance, according to the instance statistic-based similarity measure employed by MIPLIB 2017  |
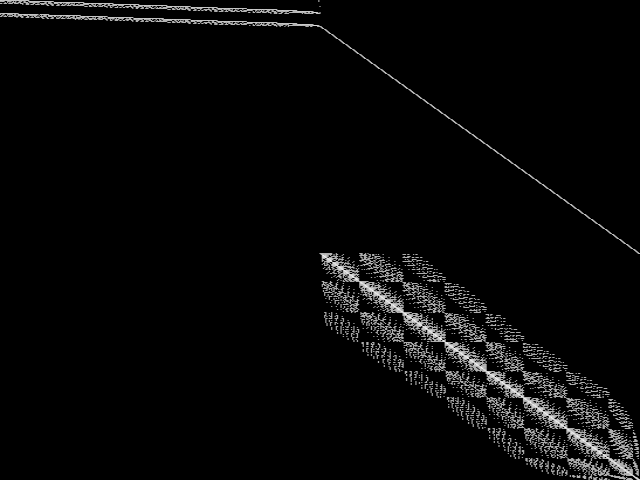
Decomposed
These decomposed images were created by GCG.
|
 |
 |
 |
 |
 |
| Name | neos-4393408-tinui [MIPLIB] | neos-4333464-siret [MIPLIB] | ns1856153 [MIPLIB] | neos-4355351-swalm [MIPLIB] | neos-4387871-tavua* [MIPLIB] | |
|
Rank / ISS
The image-based structural similarity (ISS) metric measures the Euclidean distance between the image-based feature vectors for the query instance and all model groups. A smaller ISS value indicates greater similarity.
|
1 / 0.819 | 129 / 1.470 | 564 / 1.727 | 729 / 1.857 | 1* / 0.819* | |
|
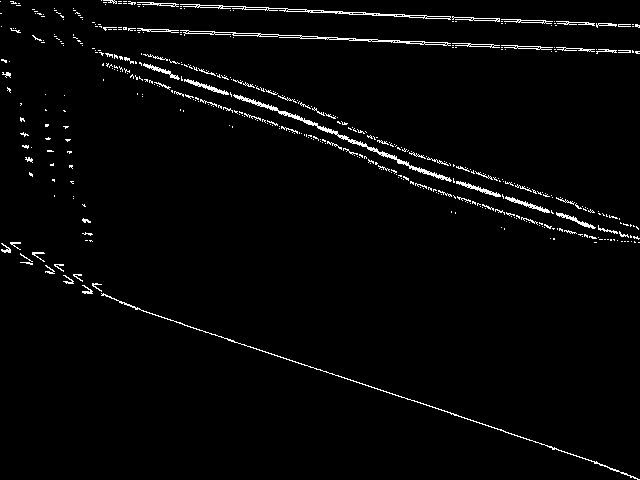
Raw
These images represent the CCM images in their raw forms (before any decomposition was applied) for the MIPLIB top 5.
|
 |
 |
 |
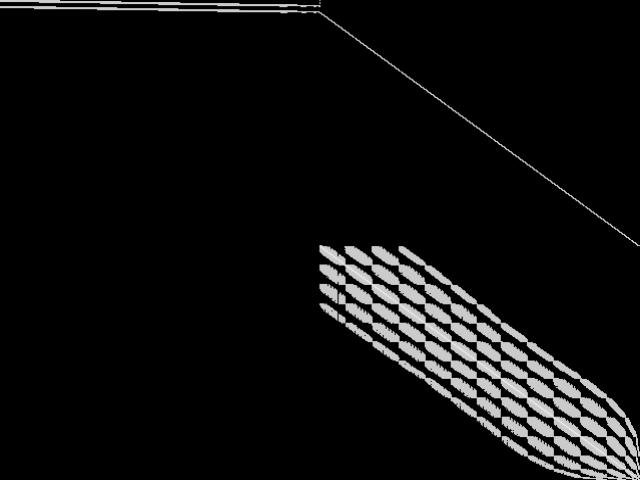 |
 |
Instance Summary
The table below contains summary information for neos-4343293-stony, the five most similar instances to neos-4343293-stony according to the MIC, and the five most similar instances to neos-4343293-stony according to MIPLIB 2017.
| INSTANCE | SUBMITTER | DESCRIPTION | ISS | RANK | |
|---|---|---|---|---|---|
| Parent Instance | neos-4343293-stony [MIPLIB] | Jeff Linderoth | (None provided) | 0.000000 | - |
| MIC Top 5 | neos-4393408-tinui [MIPLIB] | Jeff Linderoth | (None provided) | 0.819130 | 1 |
| proteindesign122trx11p8 [MIPLIB] | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 0.991427 | 2 | |
| proteindesign121pgb11p9 [MIPLIB] | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 0.992492 | 3 | |
| proteindesign121hz512p9 [MIPLIB] | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 0.996125 | 4 | |
| supportcase43 [MIPLIB] | Domenico Salvagnin | Instance coming from IBM developerWorks forum with unknown application. | 1.009362 | 5 | |
| MIPLIB Top 5 | neos-4393408-tinui [MIPLIB] | Jeff Linderoth | (None provided) | 0.819130 | 1 |
| neos-4333464-siret [MIPLIB] | Jeff Linderoth | (None provided) | 1.470343 | 129 | |
| ns1856153 [MIPLIB] | NEOS Server Submission | Sensor placement problem | 1.726844 | 564 | |
| neos-4355351-swalm [MIPLIB] | Jeff Linderoth | (None provided) | 1.856955 | 729 | |
| neos-4387871-tavua* [MIPLIB] | Jeff Linderoth | (None provided) | 0.819130* | 1* |
neos-4343293-stony: Instance-to-Model Comparison Results
| Model Group Assignment from MIPLIB: | neos-pseudoapplication-58 |
| Assigned Model Group Rank/ISS in the MIC: | 15 / 2.341 |
MIC Top 5 Model Groups
These are the 5 model group composite (MGC) images that are most similar to the decomposed CCM image for the query instance, according to the ISS metric.  |
These are model group composite (MGC) images for the MIC top 5 model groups.
|
 |
 |
 |
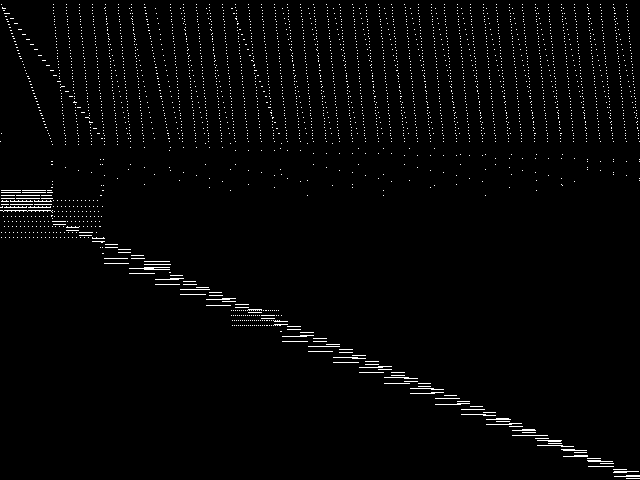 |
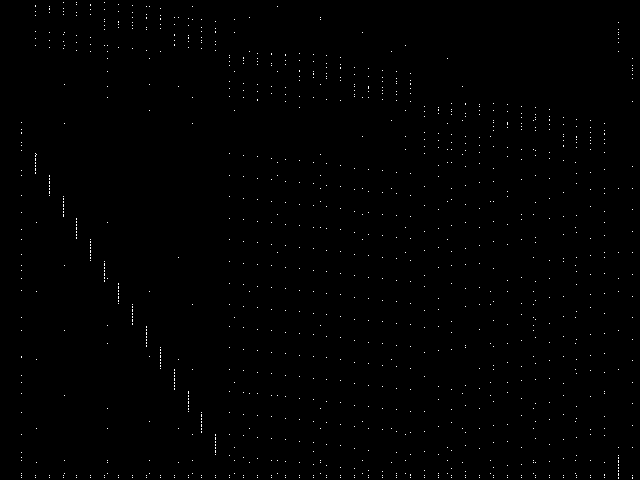 |
| Name | proteindesign | scp | neos-pseudoapplication-21 | fhnw-sq | stein | |
|
Rank / ISS
The image-based structural similarity (ISS) metric measures the Euclidean distance between the image-based feature vectors for the query instance and all other instances. A smaller ISS value indicates greater similarity.
|
1 / 1.449 | 2 / 2.100 | 3 / 2.178 | 4 / 2.181 | 5 / 2.209 |
Model Group Summary
The table below contains summary information for the five most similar model groups to neos-4343293-stony according to the MIC.
| MODEL GROUP | SUBMITTER | DESCRIPTION | ISS | RANK | |
|---|---|---|---|---|---|
| MIC Top 5 | proteindesign | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 1.449479 | 1 |
| scp | Shunji Umetani | This is a random test model generator for SCP using the scheme of the following paper, namely the column cost c[j] are integer randomly generated from [1,100]; every column covers at least one row; and every row is covered by at least two columns. see reference: E. Balas and A. Ho, Set covering algorithms using cutting planes, heuristics, and subgradient optimization: A computational study, Mathematical Programming, 12 (1980), 37-60. We have newly generated Classes I-N with the following parameter values, where each class has five models. We have also generated reduced models by a standard pricing method in the following paper: S. Umetani and M. Yagiura, Relaxation heuristics for the set covering problem, Journal of the Operations Research Society of Japan, 50 (2007), 350-375. You can obtain the model generator program from the following web site. https://sites.google.com/site/shunjiumetani/benchmark | 2.099669 | 2 | |
| neos-pseudoapplication-21 | NEOS Server Submission | Imported from the MIPLIB2010 submissions. | 2.177757 | 3 | |
| fhnw-sq | Simon Felix | Combinatorial toy fesability problem: Magic square. Models 1 & 2 are feasible, model 3 is unknown. | 2.180786 | 4 | |
| stein | MIPLIB submission pool | Imported from the MIPLIB2010 submissions. | 2.208937 | 5 |

