×
![]()
supportcase43: Instance-to-Instance Comparison Results
| Type: | Instance |
| Submitter: | Domenico Salvagnin |
| Description: | Instance coming from IBM developerWorks forum with unknown application. |
| MIPLIB Entry |
Parent Instance (supportcase43)
All other instances below were be compared against this "query" instance.  |
 |
 |
 |
 |
 |
|
Raw
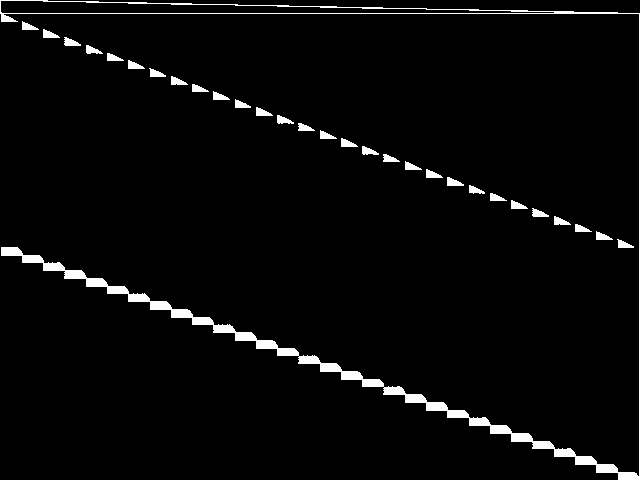
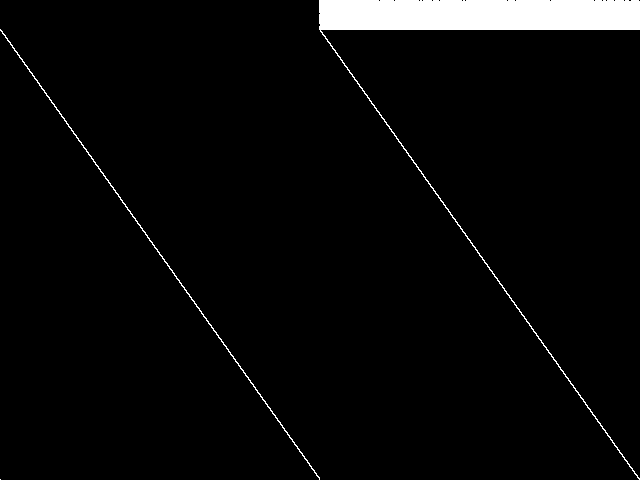
This is the CCM image before the decomposition procedure has been applied.
|
Decomposed
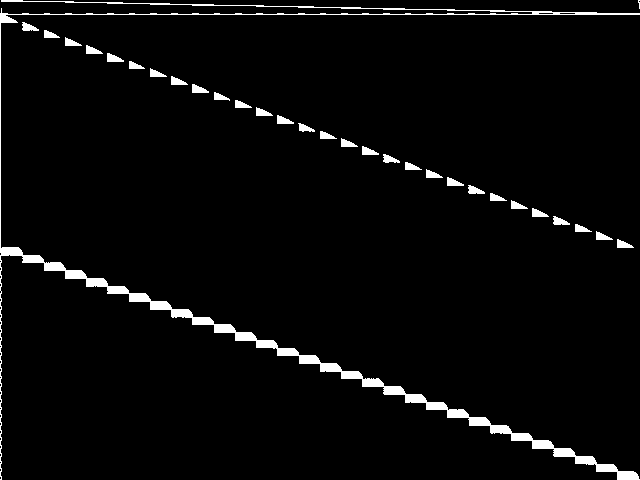
This is the CCM image after a decomposition procedure has been applied. This is the image used by the MIC's image-based comparisons for this query instance.
|
Composite of MIC Top 5
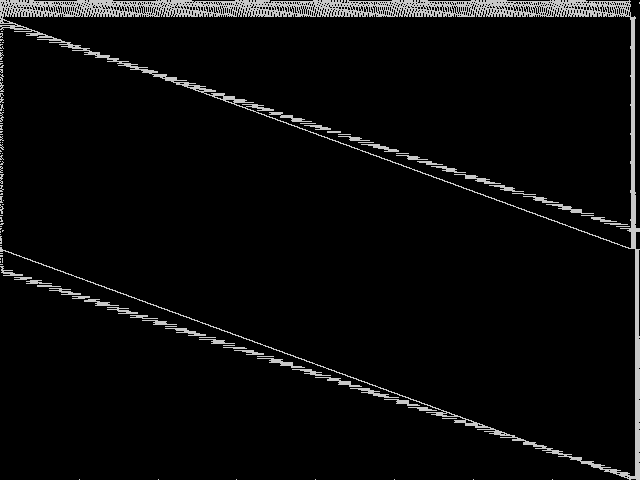
Composite of the five decomposed CCM images from the MIC Top 5.
|
Composite of MIPLIB Top 5
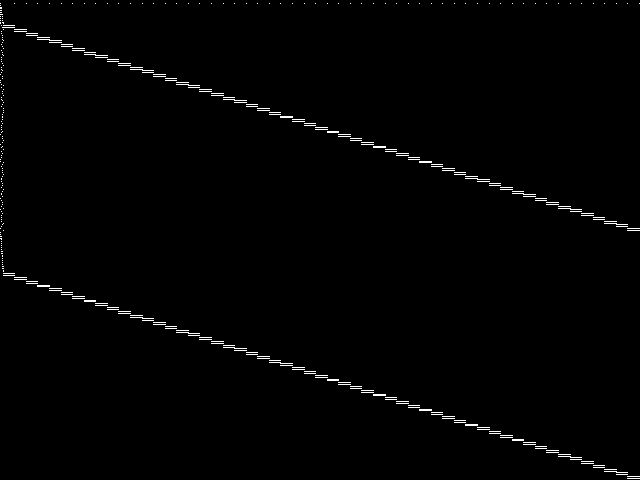
Composite of the five decomposed CCM images from the MIPLIB Top 5.
|
Model Group Composite Image
Composite of the decomposed CCM images for every instance in the same model group as this query.
|
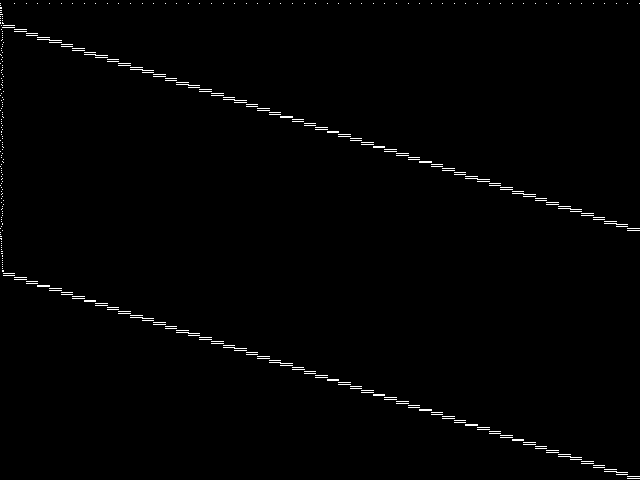
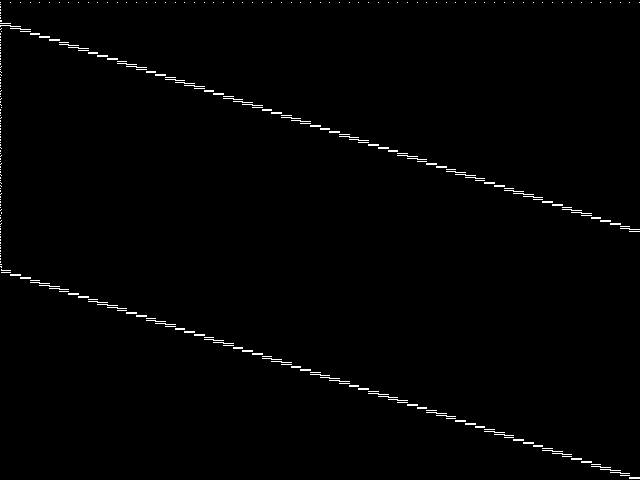
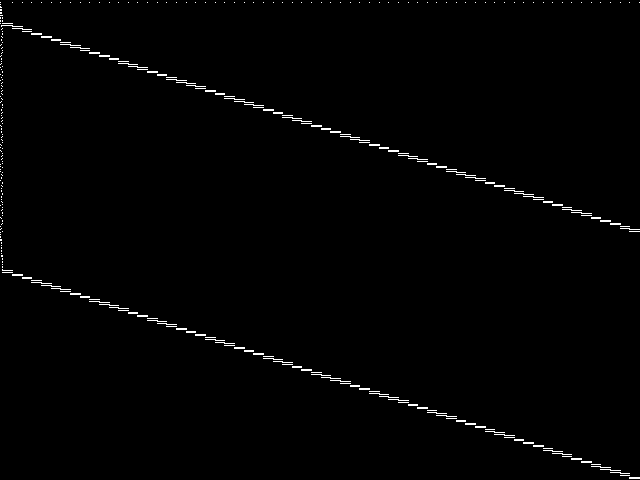
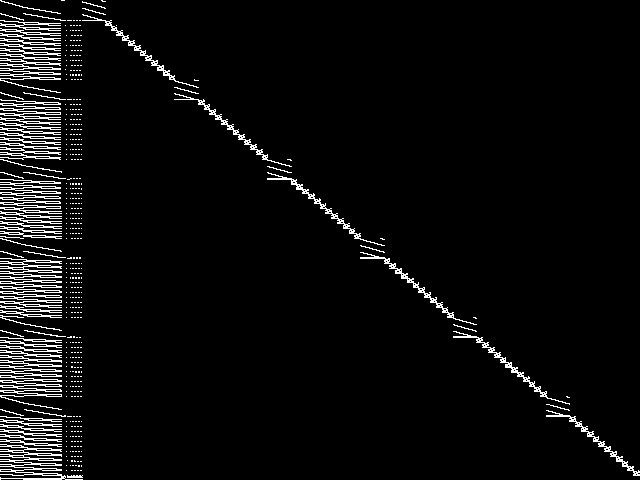
MIC Top 5 Instances
These are the 5 decomposed CCM images that are most similar to decomposed CCM image for the the query instance, according to the ISS metric.  |
Decomposed
These decomposed images were created by GCG.
|
 |
 |
 |
 |
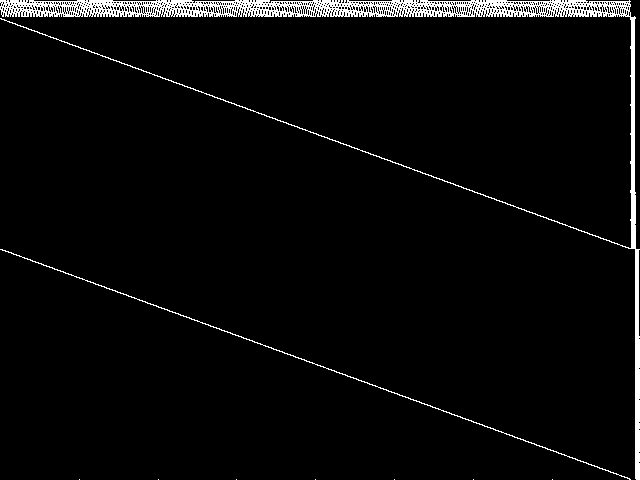 |
| Name | proteindesign122trx11p8 [MIPLIB] | proteindesign121pgb11p9 [MIPLIB] | proteindesign121hz512p19 [MIPLIB] | proteindesign121hz512p9 [MIPLIB] | ns930473 [MIPLIB] | |
|
Rank / ISS
The image-based structural similarity (ISS) metric measures the Euclidean distance between the image-based feature vectors for the query instance and all other instances. A smaller ISS value indicates greater similarity.
|
1 / 0.701 | 2 / 0.705 | 3 / 0.708 | 4 / 0.712 | 5 / 0.752 | |
|
Raw
These images represent the CCM images in their raw forms (before any decomposition was applied) for the MIC top 5.
|
 |
 |
 |
 |
 |
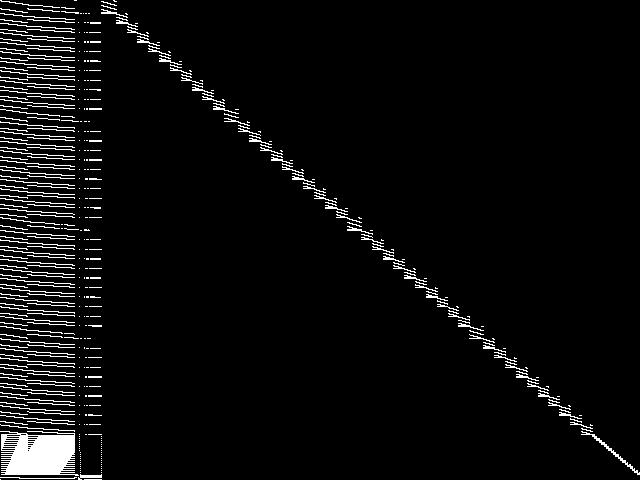
MIPLIB Top 5 Instances
These are the 5 instances that are most closely related to the query instance, according to the instance statistic-based similarity measure employed by MIPLIB 2017  |
Decomposed
These decomposed images were created by GCG.
|
 |
 |
 |
 |
 |
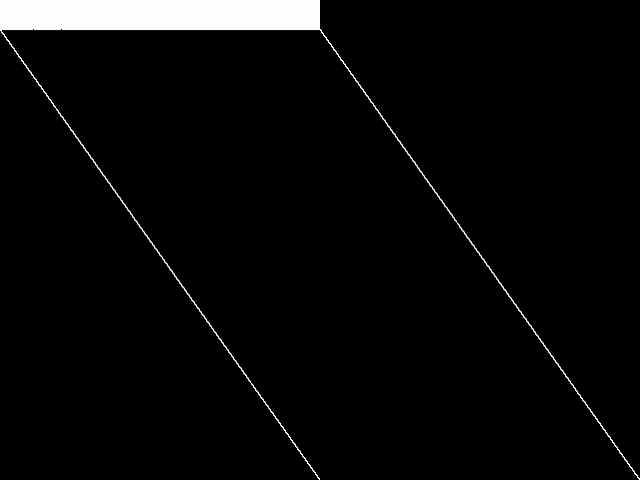
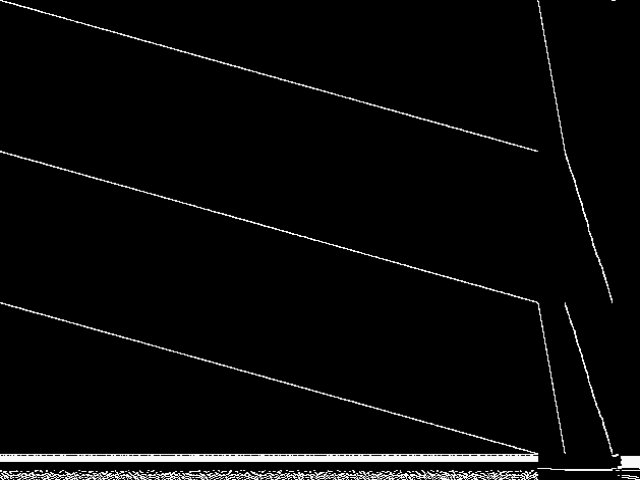
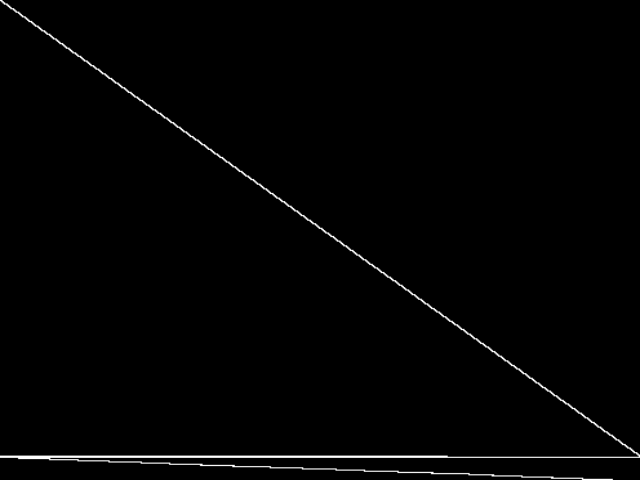
| Name | neos-872648 [MIPLIB] | neos-873061 [MIPLIB] | neos-3755335-nizao [MIPLIB] | neos-3759587-noosa [MIPLIB] | neos-872648** [MIPLIB] | |
|
Rank / ISS
The image-based structural similarity (ISS) metric measures the Euclidean distance between the image-based feature vectors for the query instance and all model groups. A smaller ISS value indicates greater similarity.
|
190 / 1.431 | 194 / 1.433 | 785 / 1.956 | 800 / 2.002 | N.A.** / N.A.** | |
|
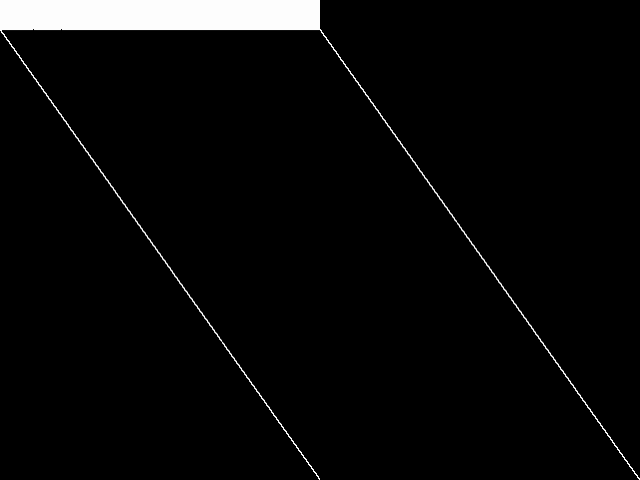
Raw
These images represent the CCM images in their raw forms (before any decomposition was applied) for the MIPLIB top 5.
|
 |
 |
 |
 |
 |
Instance Summary
The table below contains summary information for supportcase43, the five most similar instances to supportcase43 according to the MIC, and the five most similar instances to supportcase43 according to MIPLIB 2017.
| INSTANCE | SUBMITTER | DESCRIPTION | ISS | RANK | |
|---|---|---|---|---|---|
| Parent Instance | supportcase43 [MIPLIB] | Domenico Salvagnin | Instance coming from IBM developerWorks forum with unknown application. | 0.000000 | - |
| MIC Top 5 | proteindesign122trx11p8 [MIPLIB] | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 0.701222 | 1 |
| proteindesign121pgb11p9 [MIPLIB] | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 0.705013 | 2 | |
| proteindesign121hz512p19 [MIPLIB] | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 0.707895 | 3 | |
| proteindesign121hz512p9 [MIPLIB] | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 0.711647 | 4 | |
| ns930473 [MIPLIB] | NEOS Server Submission | Instance coming from the NEOS Server with unknown application | 0.751606 | 5 | |
| MIPLIB Top 5 | neos-872648 [MIPLIB] | NEOS Server Submission | Imported from the MIPLIB2010 submissions. | 1.430623 | 190 |
| neos-873061 [MIPLIB] | NEOS Server Submission | Imported from the MIPLIB2010 submissions. | 1.433202 | 194 | |
| neos-3755335-nizao [MIPLIB] | Jeff Linderoth | (None provided) | 1.955940 | 785 | |
| neos-3759587-noosa [MIPLIB] | Jeff Linderoth | (None provided) | 2.001814 | 800 | |
| neos-872648** [MIPLIB] | Armin Fuegenschuh | Packing of paths, multicommodity flow formulation. Solved in December 2013 by Gurobi development version. | N.A.** | N.A.** |
supportcase43: Instance-to-Model Comparison Results
| Model Group Assignment from MIPLIB: | no model group assignment |
| Assigned Model Group Rank/ISS in the MIC: | N.A. / N.A. |
MIC Top 5 Model Groups
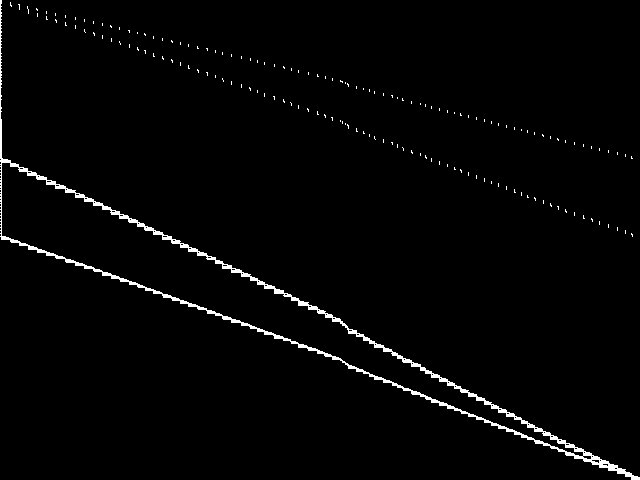
These are the 5 model group composite (MGC) images that are most similar to the decomposed CCM image for the query instance, according to the ISS metric.  |
These are model group composite (MGC) images for the MIC top 5 model groups.
|
 |
 |
 |
 |
 |
| Name | proteindesign | scp | neos-pseudoapplication-95 | neos-pseudoapplication-74 | neos-pseudoapplication-77 | |
|
Rank / ISS
The image-based structural similarity (ISS) metric measures the Euclidean distance between the image-based feature vectors for the query instance and all other instances. A smaller ISS value indicates greater similarity.
|
1 / 1.153 | 2 / 1.733 | 3 / 1.858 | 4 / 1.870 | 5 / 1.882 |
Model Group Summary
The table below contains summary information for the five most similar model groups to supportcase43 according to the MIC.
| MODEL GROUP | SUBMITTER | DESCRIPTION | ISS | RANK | |
|---|---|---|---|---|---|
| MIC Top 5 | proteindesign | Gleb Belov | Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don't know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38 | 1.153012 | 1 |
| scp | Shunji Umetani | This is a random test model generator for SCP using the scheme of the following paper, namely the column cost c[j] are integer randomly generated from [1,100]; every column covers at least one row; and every row is covered by at least two columns. see reference: E. Balas and A. Ho, Set covering algorithms using cutting planes, heuristics, and subgradient optimization: A computational study, Mathematical Programming, 12 (1980), 37-60. We have newly generated Classes I-N with the following parameter values, where each class has five models. We have also generated reduced models by a standard pricing method in the following paper: S. Umetani and M. Yagiura, Relaxation heuristics for the set covering problem, Journal of the Operations Research Society of Japan, 50 (2007), 350-375. You can obtain the model generator program from the following web site. https://sites.google.com/site/shunjiumetani/benchmark | 1.733060 | 2 | |
| neos-pseudoapplication-95 | NEOS Server Submission | Imported from the MIPLIB2010 submissions. | 1.858205 | 3 | |
| neos-pseudoapplication-74 | Jeff Linderoth | (None provided) | 1.870368 | 4 | |
| neos-pseudoapplication-77 | Jeff Linderoth | (None provided) | 1.882389 | 5 |

