×
![]()
fastxgemm
| Type: | Model Group |
| Submitter: | Laurent Sorber |
| Description: | Naive multiplication of two N by N matrices requires N^3 scalar multiplications. For N=2, Strassen showed that it could be done in only R=7 < 8=N^3 multiplications. For N=3, it is known that 19 <= R <= 23, and for N=4 it is known that 34 <= R <= 49. This repository contains code that generates a mixed-integer linear program (MILP) formulation of the fast matrix multiplication problem for finding solutions with R < N^3 and proving that they are optimal. For a more detailed description, see the accompanying manuscript. |
Parent Model Group (fastxgemm)
All other model groups below were be compared against this "query" model group.  |
 |
|
Model Group Composite (MGC) image
Composite of the decomposed CCM images for every instance in the query model group.
|
Component Instances (Decomposed)
These are the decomposed CCM images for each instance in the query model group.  |
These are component instance images.
|
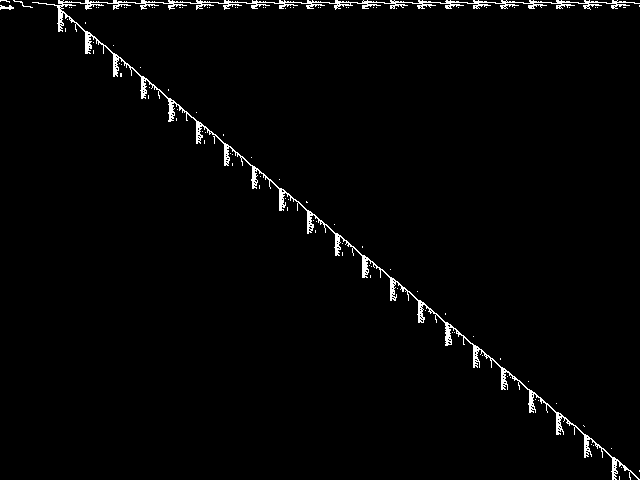 |
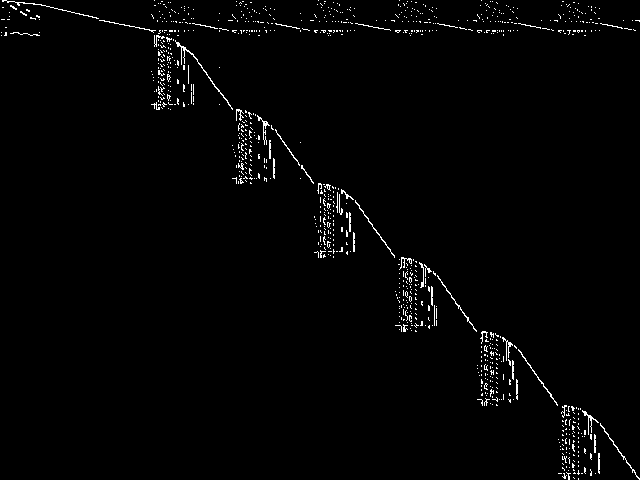 |
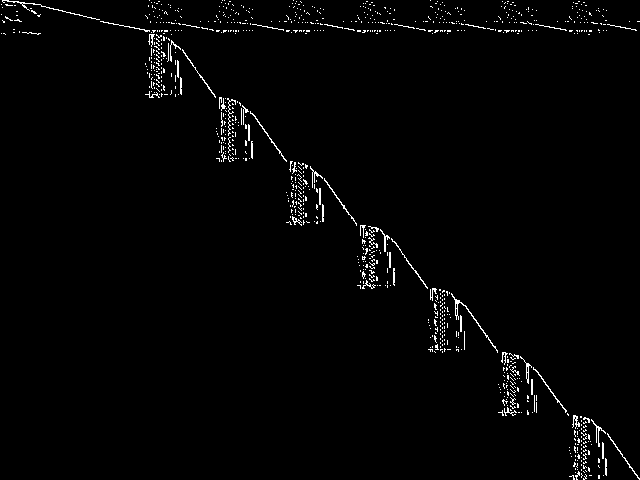 |
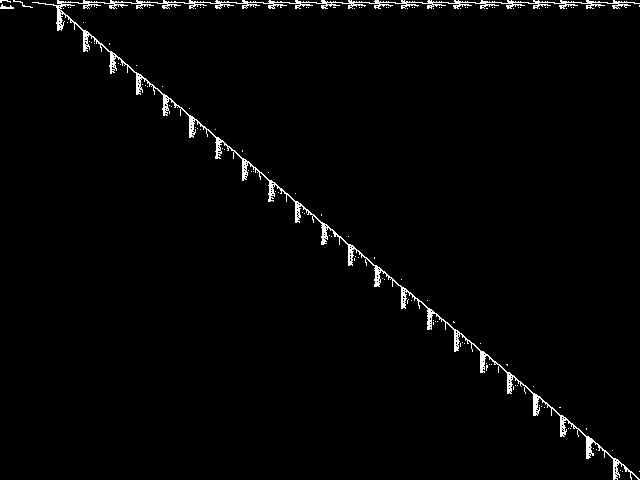 |
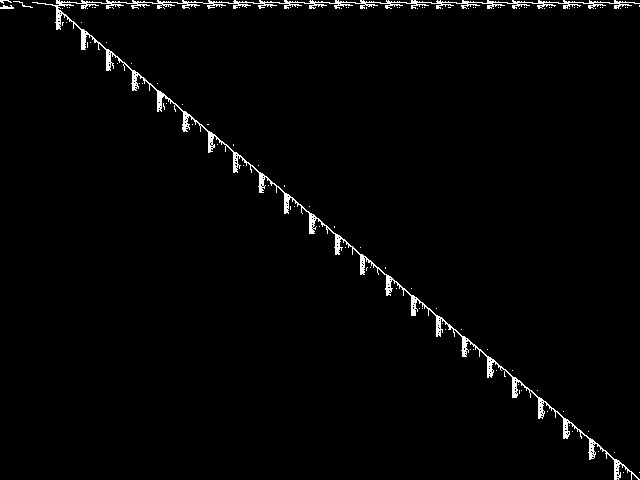 |
| Name | fastxgemm-n3r21s3t6 | fastxgemm-n2r6s0t2 | fastxgemm-n2r7s4t1 | fastxgemm-n3r22s4t6 | fastxgemm-n3r23s5t6 |
MIC Top 5 Model Groups
These are the 5 MGC images that are most similar to the MGC image for the query model group, according to the ISS metric.  |
FIXME - These are model group composite images.
|
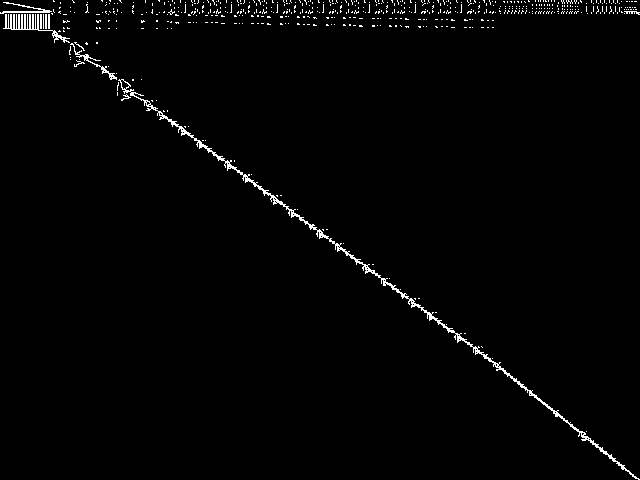 |
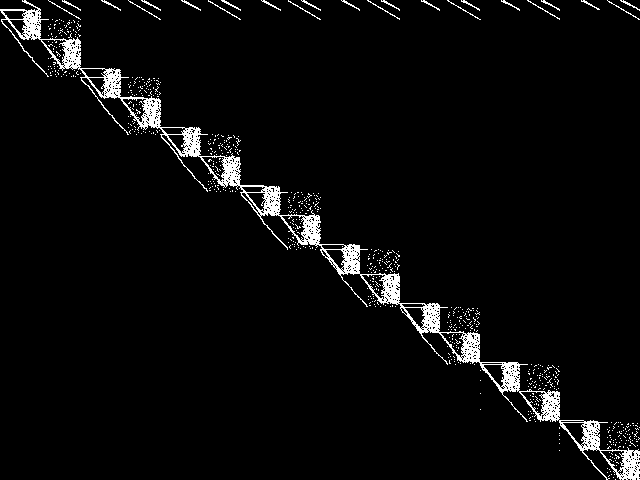 |
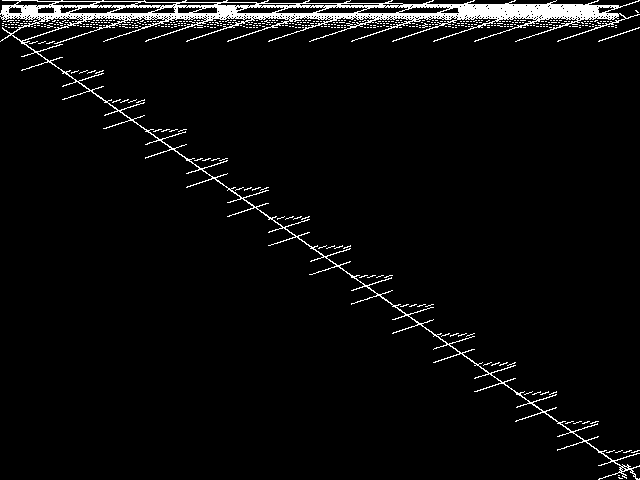 |
 |
 |
| Name | mapping | cvs | neos-pseudoapplication-86 | aflow | graphs | |
|
Rank / ISS
The image-based structural similarity (ISS) metric measures the Euclidean distance between the image-based feature vectors for the query model group and all other model groups. A smaller ISS value indicates greater similarity.
|
1 / 1.464 | 2 / 1.545 | 3 / 1.608 | 4 / 1.629 | 5 / 1.651 |
Model Group Summary
The table below contains summary information for fastxgemm, and for the five most similar model groups to fastxgemm according to the MIC.
| MODEL GROUP | SUBMITTER | DESCRIPTION | ISS | RANK | |
|---|---|---|---|---|---|
| Parent Model Group | fastxgemm | Laurent Sorber | Naive multiplication of two N by N matrices requires N^3 scalar multiplications. For N=2, Strassen showed that it could be done in only R=7 < 8=N^3 multiplications. For N=3, it is known that 19 <= R <= 23, and for N=4 it is known that 34 <= R <= 49. This repository contains code that generates a mixed-integer linear program (MILP) formulation of the fast matrix multiplication problem for finding solutions with R < N^3 and proving that they are optimal. For a more detailed description, see the accompanying manuscript. | 0.000000 | - |
| MIC Top 5 | mapping | Gleb Belov | These are the models from MiniZinc Challenges 2012-2016 (see www.minizinc.org), compiled for MIP WITH INDICATOR CONSTRAINTS using the develop branch of MiniZinc and CPLEX 12.7.1 on 30 April 2017. Thus, these models can only be handled by solvers accepting indicator constraints. For models compiled with big-M/domain decomposition only, see my previous submission to MIPLIB.To recompile, create a directory MODELS, a list lst12_16.txt of the models with full paths to mzn/dzn files of each model per line, and say$> ~/install/libmzn/tests/benchmarking/mzn-test.py -l ../lst12_16.txt -slvPrf MZN-CPLEX -debug 1 -addOption "-timeout 3 -D fIndConstr=true -D fMIPdomains=false" -useJoinedName "-writeModel MODELS_IND/%s.mps" Alternatively, you can compile individual model as follows: $> mzn-cplex -v -s -G linear -output-time ../challenge_2012_2016/mznc2016_probs/zephyrus/zephyrus.mzn ../challenge_2012_2016/mznc2016_p/zephyrus/14__8__6__3.dzn -a -timeout 3 -D fIndConstr=true -D fMIPdomains=false -writeModel MODELS_IND/challenge_2012_2016mznc2016_probszephyruszephyrusmzn-challenge_2012_2016mznc2016_probszephyrus14__8__6__3dzn.mps | 1.464023 | 1 |
| cvs | Michael Bastubbe | Capacitated vertex separator problem on randomly generated hypergraph with 128 vertices and 89 hyperedges in at most 16 components each including at most 8 vertices. solved with default GCG/Soplex in about 2000 seconds. | 1.545193 | 2 | |
| neos-pseudoapplication-86 | Jeff Linderoth | (None provided) | 1.607837 | 3 | |
| aflow | T. Achterberg | Arborescence flow problem on a graph with 40 nodes and edge density 0.9 | 1.628888 | 4 | |
| graphs | Michael Bastubbe | Packing Cuts in Undirected Graphs. Models are described in 4.1. | 1.651261 | 5 |

